The following instructions assume that you have installed SPECFEM3D and familiarized yourself with how you will run the package based on your computer configuration, as detailed in the SPECFEM3D User manual (Chapter 2 provides installation help). Additionally, we will make use of an external, hexahedral mesher CUBIT. Please make sure you have these packages installed on your system.
The example is distributed with the package under the examples/ directory. However, you might need to edit these example scripts slightly to launch them on your system.
Homogeneous halfspace
This is a step-by-step tutorial how to create a mesh for a homogeneous halfspace, export it into a SPECFEM3D file format and run the mesh partitioning and database generation.
Meshing
In the following, we will run a python script within CUBIT to create the needed mesh files for the partitioner.
- change your working directory to the example folder:
[]$ cd SPECFEM3D/examples/homogeneous_halfspace
- start the graphical user interface (GUI) of CUBIT:
[homogeneous_halfspace]$ claro
- run the python file “block_mesh.py”:
use the (Play Journal File) button and open the file 'block_mesh.py'

- in case the script executed successfully, it should create you a single volume with a regular mesh
 check that the script created a new folder “MESH/” containing the files:
check that the script created a new folder “MESH/” containing the files:
[homogeneous_halfspace]$ ls -1 MESH/ absorbing_surface_file_bottom absorbing_surface_file_xmax absorbing_surface_file_xmin absorbing_surface_file_ymax absorbing_surface_file_ymin free_surface_file materials_file mesh_file nodes_coords_file nummaterial_velocity_file
you can also check if the export was successful by examining the output in the Command line window of CUBIT. The sensitive part should look like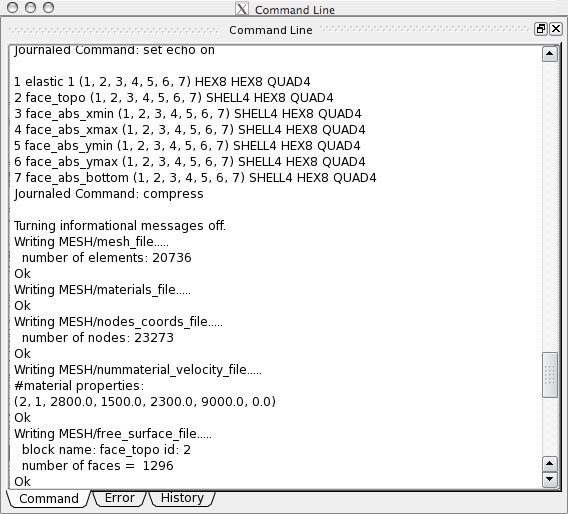
- optionally, you could modify the material properties of the halfspace, by going to change the block properties
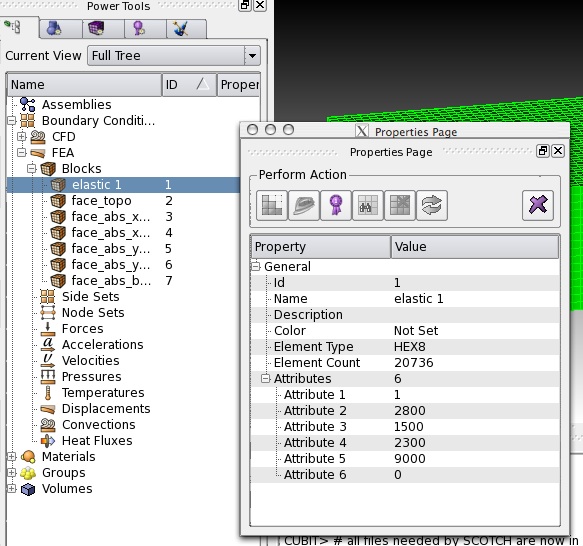 the following table applies for elastic material properties:
the following table applies for elastic material properties:
| Name | block name must start with “elastic” for elastic materials followed by a unique identifier material_id |
| Attribute 1 | Material ID |
| Attribute 2 | P-wave speed |
| Attribute 3 | S-wave speed |
| Attribute 4 | Density |
| Attribute 5 | Quality factor |
| Attribute 6 | Anisotropy flag |
Once you changed the properties, you will have to re-export the mesh. This can be done, using the script run_cubit2specfem3d.py:
use the (Play Journal File) button and open the file "run_cubit2specfem3d.py"
This should refresh the files in directory “MESH/”.
Setting up example folder for simulations
We will set up the example folder for simulation runs:
* databases directory: create a directory in_out_files/DATABASES_MPI/ into which you will put the mesh partitions:
[]$ cd SPECFEM3D/examples/homogeneous_halfspace [homogeneous_halfspace]$ mkdir -p in_out_files/DATABASES_MPI
* parameter files: make sure you have the parameter files in a local directory in_data_files/ for the example:
[homogeneous_halfspace]$ ls -1 in_data_files/ Par_file CMTSOLUTION STATIONS
these files should already be provided in the example folder.
* executables: compile the executables in the root directory:
[]$ cd SPECFEM3D/ [SPECFEM3D]$ make
in case the compilation was successful, it will create the executables xdecompose_mesh_SCOTCH, xgenerate_databases and xspecfem3D in the bin/ directory
and create a local directory bin/ to link the executables from the root directory:
[]$ cd SPECFEM3D/examples/homogeneous_halfspace [homogeneous_halfspace]$ mkdir -p bin [homogeneous_halfspace]$ cd bin/ [bin]$ ln -s ../../../bin/xdecompose_mesh_SCOTCH [bin]$ ln -s ../../../bin/xgenerate_databases [bin]$ ln -s ../../../bin/xspecfem3D
All these steps and the following decomposition, database generation and solver run are put in a process.sh bash script file in the example folder. You can simply run the script:
[homogeneous_halfspace]$ ./process.sh
to do the setup and following steps for you. Please modify and adapt the script to your needs.
Mesh partitioning
In this example, we will partition the mesh for 4 CPU cores.
run the mesh partitioner:
[homogeneous_halfspace]$ ./bin/xdecompose_mesh_SCOTCH 4 MESH/ in_out_files/DATABASES_MPI/
check that this created the mesh partitions:
[homogeneous_halfspace]$ ls -1 in_out_files/DATABASES_MPI/ proc000000_Database proc000001_Database proc000002_Database proc000003_Database
You are done.
Database generation
Next, you will need to create the mesh databases.
- in case you can run parallel programs on your desktop (needs an MPI installation), you can run the executable like:
[homogeneous_halfspace]$ cd bin/ [bin]$ mpirun -np 4 ./xgenerate_databases
otherwise, you will need to modify and adapt one of the cluster scripts provided in theSPECFEM3D/utils/Cluster/directory. - check the output file
in_out_files/OUTPUT_FILES/output_mesher.txtto see if the databases were generated successfully. The output file contains the suggested time step for your mesh:Verification of simulation parameters ... Minimum period resolved = 2.115564 **Maximum suggested time step = 6.8863556E-02**
Forward simulation
To run a forward simulation, do the following:
- make sure, you have the parameter files in the directory
in_data_files/. Most parameters in thePar_fileshould be set before running the database generation. The following may be changed after runningxgenerate_databases:# forward or adjoint simulation SIMULATION_TYPE = 1 # 1 = forward, 2 = adjoint, 3 = both simultaneously NOISE_TOMOGRAPHY = 0 # 0 = earthquake simulation, 1/2/3 = three steps in noise simulation SAVE_FORWARD = .false. # time step parameters NSTEP = 1000 DT = 0.05 # absorbing boundary conditions for a regional simulation ABSORBING_CONDITIONS = .false. # save AVS or OpenDX movies MOVIE_SURFACE = .false. MOVIE_VOLUME = .false. NTSTEP_BETWEEN_FRAMES = 200 CREATE_SHAKEMAP = .false. SAVE_DISPLACEMENT = .false. USE_HIGHRES_FOR_MOVIES = .false. HDUR_MOVIE = 0.0 # interval at which we output time step info and max of norm of displacement NTSTEP_BETWEEN_OUTPUT_INFO = 500 # interval in time steps for writing of seismograms NTSTEP_BETWEEN_OUTPUT_SEISMOS = 10000 # interval in time steps for reading adjoint traces NTSTEP_BETWEEN_READ_ADJSRC = 0 # 0 = read the whole adjoint sources at the same time # print source time function PRINT_SOURCE_TIME_FUNCTION = .false.
- in case you can run parallel programs on your desktop (needs an MPI installation), you can run the executable like:
[homogeneous_halfspace]$ cd bin/ [bin]$ mpirun -np 4 ./xspecfem3D
this example should take about 5 minutes to finish the simulation. - check the output file
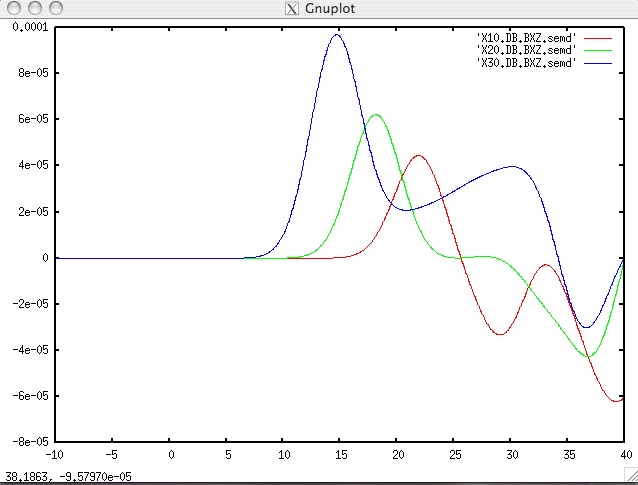
output_solver.txtin the output directoryin_out_files/OUTPUT_FILES/to see if the forward simulation was successfully finishing. the seismograms will look like this, using gnu plot:gnuplot> plot "X10.DB.BXZ.semd" w l,"X20.DB.BXZ.semd" w l,"X30.DB.BXZ.semd" w l